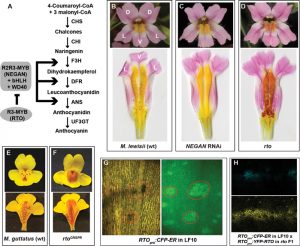
Figure 1. NEGAN and RTO constitute a two-component, activator-inhibitor RD system underlying anthocyanin spot patterning in Mimulus. (A) The anthocyanin biosynthetic pathway and its transcriptional regulators. (B-D) Anthocyanin spot patterns on the yellow background of the M. lewisii wild-type (B), NEGAN RNAi line (C), and the chemically induced rto mutant (D). (E-F) Anthocyanin patterns on nectar guides of the M. guttatus wild-type (E) and CRISPR-mediated rto mutant (F). (G) Spatial pattern of RTO promoter activity revealed by the RTOpro:CFP-ER construct. The same nectar guide area was imaged under bright light (left) and the green channel (center). (H) A transgenic plant carrying both the RTOpro:CFP-ER and the RTOpro:YFP-RTO transgenes reveal a broader spatial distribution of RTO protein (yellow) than RTO promoter activity (blue).
The emergence of complex tissue patterns from seemingly uniform, undifferentiated cells during development is an essential feature of all multicellular organisms. One of the most prominent theoretical mechanisms often invoked to explain biological pattern formation is the reaction-diffusion (RD) model, which postulates that local activation of pattern differentiation factors combined with long-range inhibition of the activity of those factors can produce dynamic, self-organizing spatial patterns. Numerous empirical and simulation studies have suggested that the RD mechanism underlies a wide range of pattern formation processes. However, we still know very little about the actual genes encoding the hypothetical activation and inhibition factors in most empirical systems, even less about the biophysical properties of these factors where candidate genes have been identified, and virtually nothing about how modulation of the properties of these activators and inhibitors affects pattern evolution in nature.
The overall objective of this project is to address these fundamental questions by elucidating the detailed genetic and developmental mechanisms of pigment pattern formation and evolution in the wildflower genus Mimulus (monkeyflowers), a system amenable to rigorous genetic analysis, developmental interrogation, and phenotypic perturbation. The work proposed here will build on our prior efforts that identified a pair of MYB proteins underlying the formation of dispersed anthocyanin pigment spots in Mimulus flowers (Figure 1; Ding et al. 2020). This MYB pair forms a local autocatalytic feedback loop and a long-range inhibitory feedback loop, fulfilling the tenets of a classical activator-inhibitor RD model. Our goals in the coming years are to:
(i) experimentally determine the biophysical properties of the activator-inhibitor pair, including their diffusion coefficients, degradation (clearance) rates, and relative activation and inhibition constants; (ii) characterize the genetic and developmental bases of pattern evolution from dispersed spots to longitudinal stripes between closely related species; and (iii) identify the key cis-regulatory elements that constitute the activator-inhibitor interacting network and test the function of this two-component, activator-inhibitor module in other tissue types and heterologous systems.
Towards these ends we will use a suite of approaches, including fluorescence imaging, genetic mapping, transgenic manipulation, and mathematical modeling. Together our efforts will provide an in-depth view of how the RD mechanism generates self-organizing spatial patterns and modulates pattern evolution in a real biological system, lending empirical support to the mathematically elegant but somewhat controversial RD model.
This project is supported by an NIH grant (R01GM140092).