This is a continuous collaboration with Dr. Toby Bradshaw, who initiated the molecular genetic analysis of floral trait variation between Mimulus lewisii and M. cardinalis in the early 1990s, and has since been leading the effort to dissect the genetic basis of floral trait divergence underlying pollinator preference, and, therefore, reproductive isolation between the two species, gene by gene, mutation by mutation.
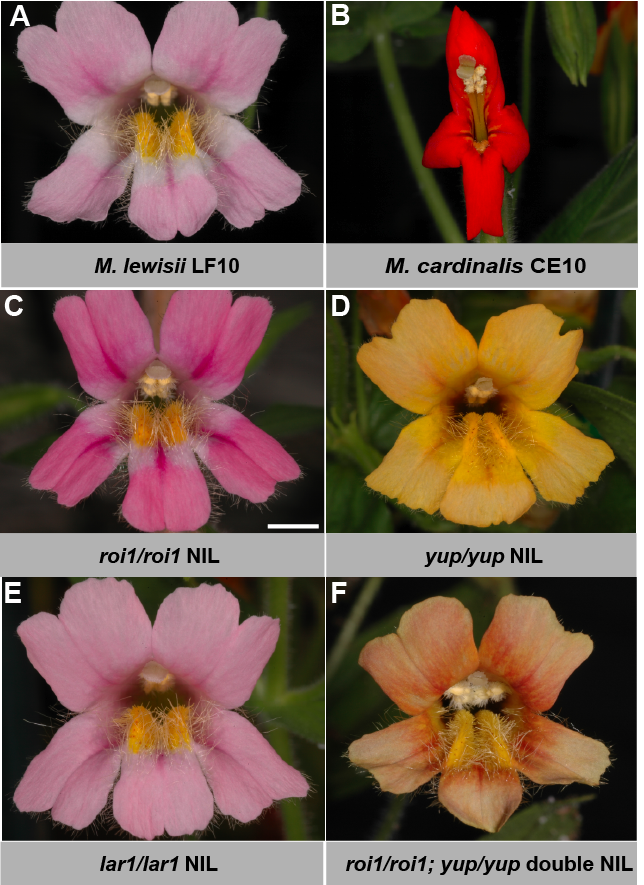
The pale pink color of M. lewisii flowers results from a low concentration of anthocyanins and the absence of carotenoids (except in the nectar guides, Figure 1A). The red color of M. cardinalis flowers (Figure 1B) is produced by a combination of high concentrations of both pigments. Both anthocyanin and carotenoid concentrations contribute to pollinator discrimination between the two species in their natural habitat (Schemske and Bradshaw 1999).
The combination of three loci explains much of the flower color difference between the two species. ROI1 accounts for the anthocyanin concentration difference; LAR1 is responsible for the presence vs. absence of the white region around the corolla throat (i.e. anthocyanin pigment patterning); YUP is responsible for the presence vs. absence of yellow carotenoids in the petal lobes.
So far we have identified ROI1 and LAR1. ROI1 is an R3–MYB gene encoding a repressor that negatively regulate anthocyanin biosynthesis (Yuan et al. 2013). ROI1 is expressed ca. 11 fold higher in M. lewisii than in M. cardinalis, leading to a darker pink phenotype when the M. lewisii allele is replaced by the M. cardinalis allele (Figure 1C). The ROI1 allelic difference is due to cis-regulatory change that remains to be identified. LAR1 encodes an R2R3-MYB TF that activates flavonol biosynthesis, which competes with the anthocyanin biosynthetic pathway for substrates and produces the spatial pattern of anthocyanin pigmentation in M. lewisii (Yuan et al. 2016). The lar1 allele in M. cardinalis is expressed ca. 40-fold lower than in M. lewisii, explaining the lack of patterning.